As part of FAIR-EASE, the Galaxy Europe Earth System subdomain
([earth
Figure 1:Earth System subdomain welcome page
It supports a wide range of scientific and operational needs, and strengthens links with European data infrastructures such as Copernicus, CMEMS and OBIS.
These tools help make environmental data more accessible and usable by:
Connecting users to major data sources through simple, reproducible workflows ;
Offering both batch processing and interactive exploration, all inside the Galaxy platform ;
Supporting training, education and open science thanks to public code, Docker images and documentation ;
Enabling long-term reusability and compatibility with EOSC and other European initiatives.
Tools integrated¶
These tools show how FAIR-EASE helps bridge the gap between data providers and users, making complex data more usable for science, policy, and operational services.
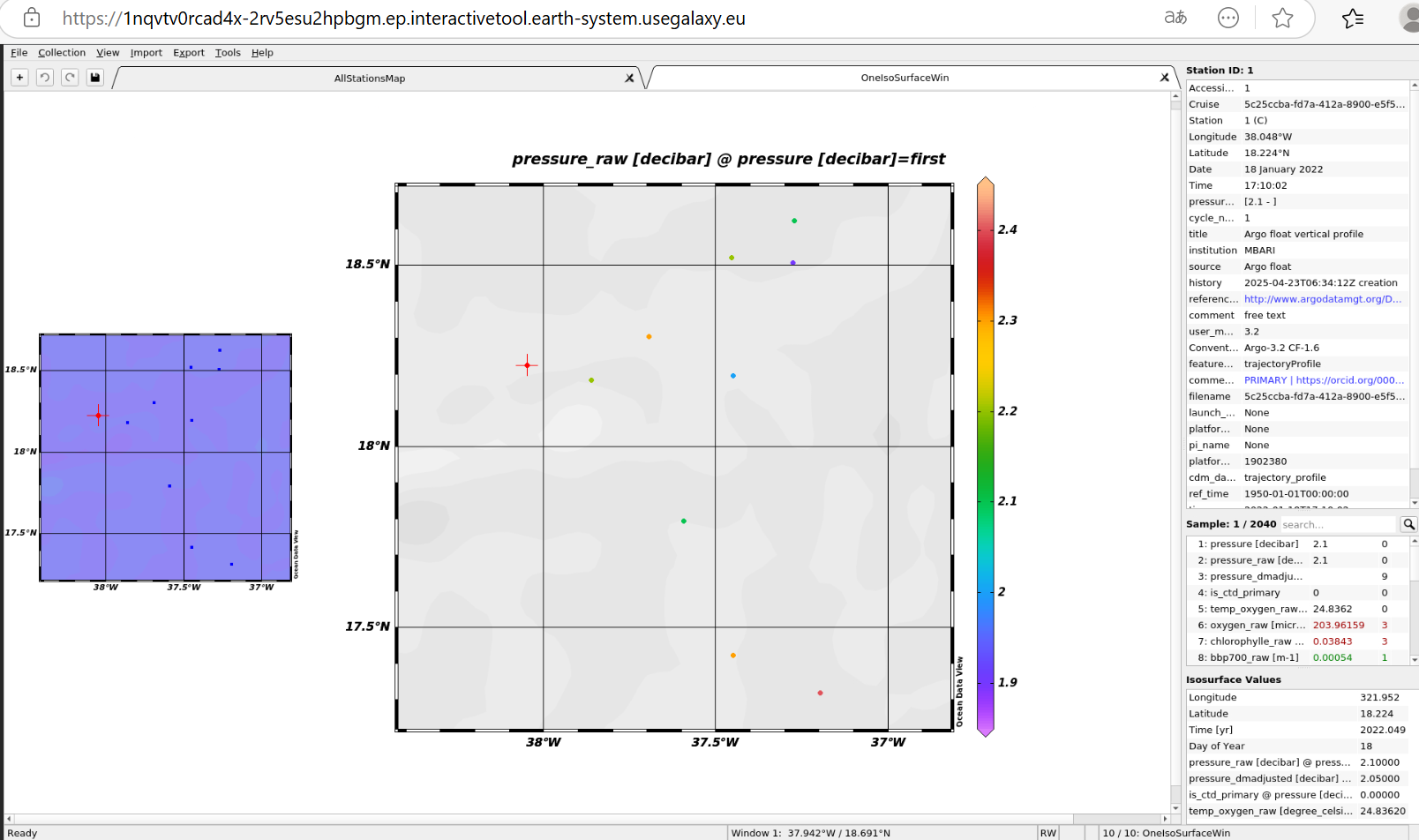
Figure 2:ODV interactive tool in Galaxy
Tools for Oceanographic Data
| Tool | Description | Links |
|---|---|---|
| Argo_getdata | Allows retrieval of Argo glider data (physical and biogeochemical). | |
| DIVA_full_analysis | Implemented as both batch and interactive tools, this module enables advanced spatial interpolation of marine data. | |
| Copernicus Marine Data Store (copernicusmarine) | Batch tool to query and download datasets from CMEMS. | |
| Ocean Data View | Used to plot geo-referenced ocean data from NetCDF and other formats. | |
| ODV collection manager (tool_odv) | Merges various datasets with a common vocabulary and creates a single generic ODV spreadsheet in an automatic way | |
| ODV history manager (tool_odv_history) | Report in the input file the ODV history including the change of QC flag | |
Canyon B (bgc_canyonb) | Robust Estimation of Open Ocean CO2 Variables and Nutrient Concentrations From T, S, and O2 Data Using Bayesian Neural Network | |
Sanntis (sanntis_marine) | The Sanntis tool identify biosynthetic gene clusters (BGCs) in genomic & metagenomic data | |
| QCV Harmonizer (harmonize_insitu_to_netcdf) | Harmonizes oceanographic biogeochemical data. |
Tools for Interactive Visualisation and Data Handling
| Tool | Description | Links |
|---|---|---|
| QGIS | Full integration of QGIS as an interactive tool in Galaxy. | |
| HoloViz Ecosystem | A set of interactive notebooks for data visualisation using Python libraries as Panel, Bokeh, Datashader, etc. | |
| STAC Browser | Access and navigation interface for STAC (SpatioTemporal Asset Catalogs). | |
| TerriaMap | Geospatial visualisation |
Access to Global and Biodiversity Data
| Tool | Description | Links |
|---|---|---|
| OBIS occurences (obis_data) | A tool to search and retrieve species occurrences from the OBIS database. | |
| Copernicus Data Space Ecosystem | Jupyter notebooks to explore Copernicus data using SentinelHub and OpenEO. | |
| Trends.Earth | Batch tool for computing land cover and degradation indicators, supporting monitoring of SDG 15.3.1. |
Workflows developed and shared¶
Several Galaxy workflows have been developed and shared as part of FAIR-EASE to support Earth system science. These include workflows to process and analyse Argo float data, extract biogeochemical variables like phosphate from large NetCDF datasets, and combine oceanographic data with tools like ODV and the Pangeo ecosystem. All workflows are openly available on WorkflowHub - [FAIR-EASE Galaxy Project on WorkflowHub] - and can be imported into Galaxy by clicking the “Run on Galaxy” button on the WorkflowHub pages. They are designed for environmental scientists working with oceanographic and Earth system data, and are compatible with the [Galaxy Europe For Earth System instance.]
In addition, these workflows were demonstrated during major scientific events such as EGU 2024, and training materials have been created to help new users. Tutorials are available through the Galaxy Training Network, showing how to run the workflows, understand the data, and use FAIR practices like RO-Crate to describe and share results.
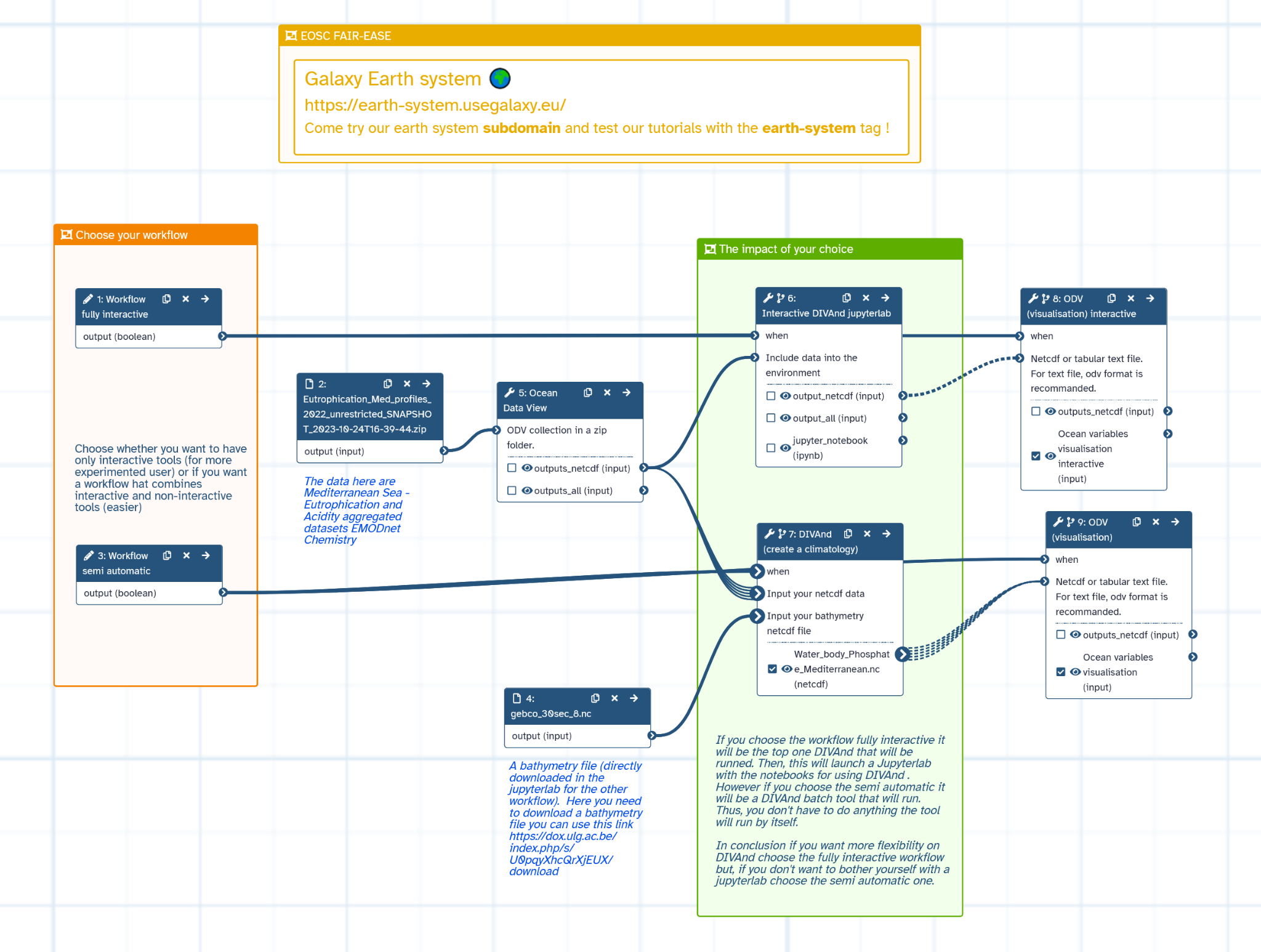
Figure 3:Water Coastal Dynamics workflow
| Workflow | Description | Access |
|---|---|---|
| Marine Omics: Biosynthetic Gene Clusters | Detects biosynthetic gene clusters in marine omics data using tools like Prodigal and SanntiS. | WorkflowHub |
| Marine Omics Visualisation (OBIS Indicators) | Converts OBIS biodiversity records into indicators such as Shannon and Simpson indices. | WorkflowHub |
| Process Argo Data with Pangeo & ODV | Processes Argo float data and visualizes oceanographic variables using Pangeo and Ocean Data View (ODV). | WorkflowHub |
| Subset Mediterranean Sea & Extract Phosphate | Subsets NetCDF data for the Mediterranean Sea and extracts phosphate levels for analysis. | WorkflowHub |
| Full Analysis of Argo Data | End-to-end workflow to analyse and visualise Argo profile datasets. | WorkflowHub |
| Argo-Glider Nitrate QCV | Qualification, Calibration and validation of Argo floats and Gliders ocean Biogeochemical Data Using Galaxy | Galaxy Earth System |
Galaxy Training Network (GTN)¶
Several high-quality, FAIR-aligned tutorials and learning pathways have been developed and published on the Galaxy Training Network (GTN). These resources are tagged with the “earth-system” label and cover a wide range of topics including marine biodiversity, oceanographic data processing, land monitoring, and FAIR metadata practices. Designed for interdisciplinary Earth and environmental scientists, they support hands-on learning with real datasets and workflows, and encourage reuse, openness, and reproducibility. All materials are licensed under CC-BY 4.0.
Figure 4:GTN example
| Title | Type | Description | Access |
|---|---|---|---|
| Getting your hands-on earth data | Learning Pathway | Introduction to accessing and analyzing ocean, land, atmosphere, biodiversity data in Galaxy. | Run Tutorial |
| OBIS Marine Indicators | Tutorial | Calculate biodiversity indices (Shannon, Simpson, ES50) from OBIS. | Run Tutorial |
| From NDVI with OpenEO to time series with Holoviews | Tutorial | Process NDVI satellite data for land degradation analysis and time-series visualization. | Run Tutorial |
| Marine Omics: Identifying BGCs | Tutorial | Detect biosynthetic gene clusters using Prodigal, InterProScan, SanntiS. | Run Tutorial |
| Ocean’s Variables Study | Tutorial | Subset Mediterranean ocean data and explore variables (e.g., phosphate). | Run Tutorial |
| Ocean Data View (ODV) | Tutorial | Visualize NetCDF-based oceanographic variables using ODV. | Run Tutorial |
| Sentinel 5P Data Visualisation | Tutorial | Explore and analyze Sentinel-5P atmosphere data interactively. | Run Tutorial |
| Analyse Argo Data | Tutorial | Process Argo float datasets with Pangeo tools and visualize using ODV | Run Tutorial |
| Make your tools available on your subdomain | Tutorial | Guide to managing tools in a Galaxy subdomain. | Run Tutorial |
| Create a subdomain for your community | Tutorial | Steps to create and administer a Galaxy subdomain for your community. | Run Tutorial |
Connecting IT resources¶
As part of FAIR-EASE, several actions were undertaken to integrate and leverage PULSAR as a distributed execution backend within Galaxy workflows. PULSAR endpoints were deployed on multiple infrastructures, including at the University of Clermont Auvergne (UCA) and the Hellenic Centre for Marine Research (HCMR) in Greece, enabling remote processing from Galaxy. A proof-of-concept with the EGI Federated Cloud was also carried out, demonstrating the ability to dynamically deploy PULSAR nodes using the EGI Infrastructure Manager. These deployments supported the execution of real-world workflows, validating the portability, scalability, and interoperability of distributed processing in an EAL platform.
Figure 5:French Galaxy Pulsar endpoint at the UCA