Last update 👈
David Palecek, May 26, 2025
MetaGOflow is a slim version of MGnify pipeline 1. Both taxonomy and functioinal annotation is done from reads, however contigs are assembled using MEGAHIT too.
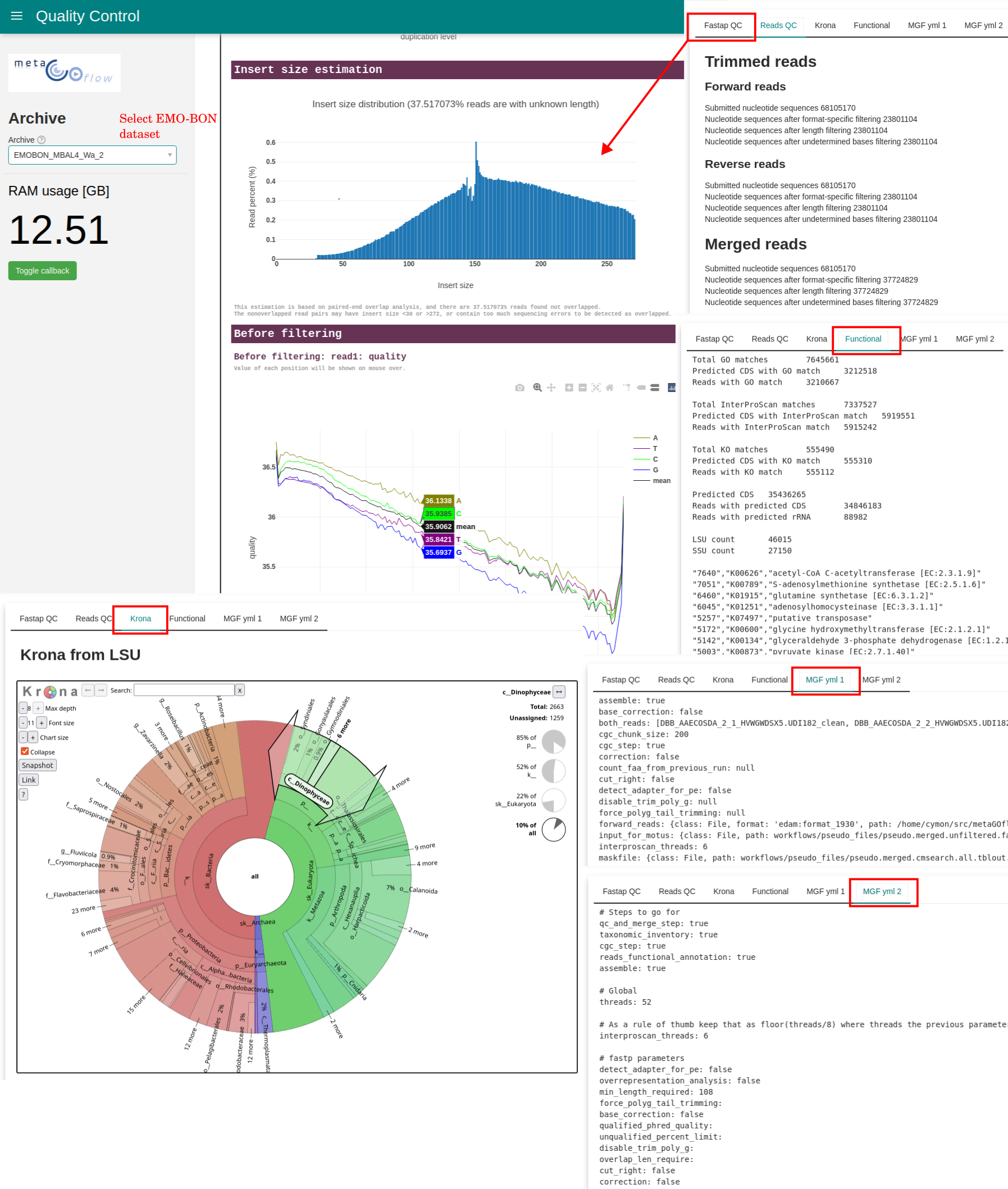
There are almost 60 output files from the metaGOflow pipeline. This dashboard provides interface to the most relevant and not too big metaGOflow pipeline outputs, including:
fastpQaulity Control report with interactive QC plots.- Reads Quality control, both
trimmedandmergedreads. - Interactive Krona plots from SSU and LSU taxonomy tables, respectively.
- Functional annotation summaries expressed in number of reads matched to respective databases.
- Zafeiropoulos, H., Beracochea, M., Ninidakis, S., Exter, K., Potirakis, A., De Moro, G., Richardson, L., Corre, E., Machado, J., Pafilis, E., Kotoulas, G., Santi, I., Finn, R. D., Cox, C. J., & Pavloudi, C. (2022). metaGOflow: a workflow for the analysis of marine Genomic Observatories shotgun metagenomics data. GigaScience, 12. 10.1093/gigascience/giad078